TRD#2: Optimizing Performance, Resolution, and Throughput of CryoTEM
At the outset of this last funding period we were at the very beginning of the “resolution revolution” that has swept cryoEM to a new and much more prominent role in structural biology. The new generation of direct detector (DD) cameras had just started to become available. We knew they had the potential to provide advantages over CCD cameras in terms of improved spatial resolution as well as the signal to noise ratio of the acquired images. Furthermore, we knew that as the data is acquired as a set of multiple image frames, each corresponding to a fraction of the total dose, the DD offered the potential of correcting for specimen drift and for calculating maps from limited dos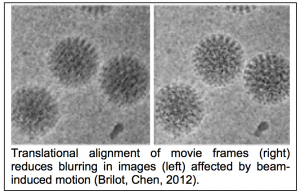 e exposures. NRAMM was fortunate to be one of the first groups to have access to a direct detector, an early version of the Direct Electron DE12. We began by integrating the camera into Leginon and characterizing its basic performance (Milazzo, Cheng, 2011) and in doing so discovered several issues with the hardware and the software that we worked closely with the company to correct. Shortly after, Niko Grigorieff arrived at NRAMM to spend a year on sabbatical. With the help of the NRAMM team he led an effort to use the movie mode of the camera to study beam induced motion (see figure 1.6) of a vitrified sample during its exposure to the electron beam (Brilot, Chen, 2012). This was one of the earliest papers to confirm that the significant blurring in recorded images indeed resulted from a drum like motion of the entire ice layer and that the specimen moves significantly more at the start of an exposure than towards its end. We immediately followed up on this work by showing that alignment and averaging of movie frames can be used to restore high-resolution detail in images affected by beam-induced motion, leading to better resolution and improved data collection efficiency (Campbell, Cheng, 2012).
e exposures. NRAMM was fortunate to be one of the first groups to have access to a direct detector, an early version of the Direct Electron DE12. We began by integrating the camera into Leginon and characterizing its basic performance (Milazzo, Cheng, 2011) and in doing so discovered several issues with the hardware and the software that we worked closely with the company to correct. Shortly after, Niko Grigorieff arrived at NRAMM to spend a year on sabbatical. With the help of the NRAMM team he led an effort to use the movie mode of the camera to study beam induced motion (see figure 1.6) of a vitrified sample during its exposure to the electron beam (Brilot, Chen, 2012). This was one of the earliest papers to confirm that the significant blurring in recorded images indeed resulted from a drum like motion of the entire ice layer and that the specimen moves significantly more at the start of an exposure than towards its end. We immediately followed up on this work by showing that alignment and averaging of movie frames can be used to restore high-resolution detail in images affected by beam-induced motion, leading to better resolution and improved data collection efficiency (Campbell, Cheng, 2012).
As a result of this early work NRAMM was able to acquire a new Gatan K2 Summit camera that provided improved performance due to its ability to count individual electrons. This camera was initially installed on a Tecnai F20, allowing us to investigate the limits of performance from these mid-range instruments (Veesler, Campbell, 2013) and show that they can be used to obtain maps of sufficient quality for de novo modeling of protein structures. Our 3.7Å map of Nudaurelia capensis ω virus (Campbell, Kearney, 2014) remains as one of the highest resolution structures ever obtained from a mid-range instrument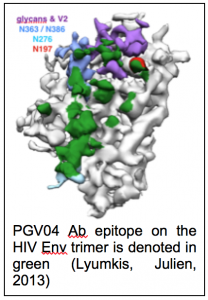 . One of our driving biological projects during this period was to obtain a structure of the HIV trimer (figure 1.7), an important target for vaccine development (Julien, Cupo, 2013; Lyumkis, Julien, 2013) and partly as a result of the success of this project we obtained funding for a new Titan Krios microscope that was installed in the first quarter of 2014. Initially, we worked with a Falcon 2 camera on loan from FEI but later swapped our K2 Summit from the F20 to the Krios. We then set out to explore the limits of performance on this very high-end combination of instruments. This work resulted in a 2.8Å map of the 20S proteasome (Campbell, Veesler, 2015), which at the time it was announced was one of the first sub 3Å single particle reconstructions available and one of the first to show that well ordered water molecules could be identified in single particle cryoEM maps. This work, and similar efforts from other labs that followed along shortly after, unequivocally demonstrated that single particle cryoEM is capable of competing with X-ray crystallography for determination of protein structures of suitable quality for rational drug design.
. One of our driving biological projects during this period was to obtain a structure of the HIV trimer (figure 1.7), an important target for vaccine development (Julien, Cupo, 2013; Lyumkis, Julien, 2013) and partly as a result of the success of this project we obtained funding for a new Titan Krios microscope that was installed in the first quarter of 2014. Initially, we worked with a Falcon 2 camera on loan from FEI but later swapped our K2 Summit from the F20 to the Krios. We then set out to explore the limits of performance on this very high-end combination of instruments. This work resulted in a 2.8Å map of the 20S proteasome (Campbell, Veesler, 2015), which at the time it was announced was one of the first sub 3Å single particle reconstructions available and one of the first to show that well ordered water molecules could be identified in single particle cryoEM maps. This work, and similar efforts from other labs that followed along shortly after, unequivocally demonstrated that single particle cryoEM is capable of competing with X-ray crystallography for determination of protein structures of suitable quality for rational drug design.
Over the last 4 years we have also expanded Leginon to support a very wide variety of microscopes (FEI Krios, Talos, Tecnai; JEOL 32200, 2200, 2100, 1230) and cameras (Gatan K2; Direct Electron DE12, DE20; FEI Falcon I, II, III as well as numerous varieties of CCD cameras). Many of these instruments required considerable effort to support due to lens hysteresis and instabilities and lack of precision in the stages. Integrating new direct detectors required optimization of motion correction algorithms specific to each camera. Numerous additional features were also added to Leginon including various controls for switching between cameras; tools for achieving coma free alignment; shutter, grid exchange and cryogen refill controls for the Titan Krios. A total of ~900 revisions were made to the Leginon base code over the past 4 years.
References
Brilot, A.F., Chen, J.Z., Cheng, A., Pan, J., Harrison, S.C., Potter, C.S., Carragher, B., Henderson, R., and Grigorieff, N. (2012). Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol 177, 630-637. Pubmed.
Campbell, M.G., Cheng, A., Brilot, A.F., Moeller, A., Lyumkis, D., Veesler, D., Pan, J., Harrison, S.C., Potter, C.S., Carragher, B., et al. (2012). Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20, 1823-1828. Pubmed.
Campbell, M.G., Kearney, B.M., Cheng, A., Potter, C.S., Johnson, J.E., Carragher, B., and Veesler, D. (2014). Near-atomic resolution reconstructions using a mid-range electron microscope operated at 200 kV. J Struct Biol 188, 183-187. Pubmed.
Campbell, M.G., Veesler, D., Cheng, A., Potter, C.S., and Carragher, B. (2015). 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife 4. Pubmed.
Julien, J.P., Cupo, A., Sok, D., Stanfield, R.L., Lyumkis, D., Deller, M.C., Klasse, P.J., Burton, D.R., Sanders, R.W., Moore, J.P., et al. (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477-1483. Pubmed.
Lyumkis, D., Julien, J.P., de Val, N., Cupo, A., Potter, C.S., Klasse, P.J., Burton, D.R., Sanders, R.W., Moore, J.P., Carragher, B., et al. (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484-1490. Pubmed.
Milazzo, A.C., Cheng, A., Moeller, A., Lyumkis, D., Jacovetty, E., Polukas, J., Ellisman, M.H., Xuong, N.H., Carragher, B., and Potter, C.S. (2011). Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. J Struct Biol 176, 404-408. Pubmed.
Veesler, D., Campbell, M.G., Cheng, A., Fu, C.Y., Murez, Z., Johnson, J.E., Potter, C.S., and Carragher, B. (2013). Maximizing the potential of electron cryomicroscopy data collected using direct detectors. J Struct Biol 184, 193-202. Pubmed.

