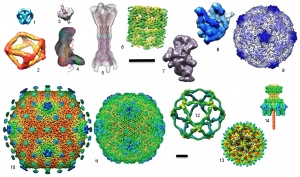
3D maps using NRAMM automation
3D maps using NRAMM automation:
3D maps of some of the macromolecular machines solved using the automated methods developed at NRAMM. Structures are: (1) 10-stranded RNA cube (Afonin et al., 2010); (2) tRNA antiprism (Severcan et al., 2010); (3) bacteriophage P22 terminase small subunit (Nemecek et al., 2008); (4) human dicer (Lau et al., 2009); (5) MacAB-TolC efflux pump (Xu et al., 2011); (6) groEL (Stagg et al., 2006; Stagg et al., 2008a); (7) 30S ribosome intermediate assembly subunits (Mulder et al., 2010); (8) 50S ribosome subunit (Voss et al., 2010); (9) Poliovirus (Hogle group); (10) SIO2 bacteriophage; (11) Lambda bacteriophage (Lander et al., 2008); (12) COPII coat cage (Stagg et al., 2007; Stagg et al., 2008b); (13) Hepatitus B virus like particles (Strable et al., 2008); (14) P22 tail machine (Lander et al., 2009). Scale bars are both 10nm; images in top section are shown at a larger scale.
Afonin, K.A., Bindewald, E., Yaghoubian, A.J., Voss, N., Jacovetty, E., Shapiro, B.A., and Jaeger, L. (2010). In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5, 676-682.
Lander, G.C., Evilevitch, A., Jeembaeva, M., Potter, C.S., Carragher, B., and Johnson, J.E. (2008). Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16, 1399-1406.
Lander, G.C., Khayat, R., Li, R., Prevelige, P.E., Potter, C.S., Carragher, B., and Johnson, J.E. (2009). The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure 17, 789-799.
Lau, P.W., Potter, C.S., Carragher, B., and MacRae, I.J. (2009). Structure of the human Dicer-TRBP complex by electron microscopy. Structure 17, 1326-1332.
Mulder, A.M., Yoshioka, C., Beck, A.H., Bunner, A.E., Milligan, R.A., Potter, C.S., Carragher, B., and Williamson, J.R. (2010). Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330, 673-677.
Nemecek, D., Lander, G.C., Johnson, J.E., Casjens, S.R., and Thomas, G.J., Jr. (2008). Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol 383, 494-501.
Severcan, I., Geary, C., Chworos, A., Voss, N., Jacovetty, E., and Jaeger, L. (2010). A polyhedron made of tRNAs. Nat Chem 2, 772-779.
Stagg, S.M., Lander, G.C., Pulokas, J., Fellmann, D., Cheng, A., Quispe, J.D., Mallick, S.P., Avila, R.M., Carragher, B., and Potter, C.S. (2006). Automated cryoEM data acquisition and analysis of 284742 particles of GroEL. J Struct Biol 155, 470-481.
Stagg, S.M., Lander, G.C., Quispe, J., Voss, N.R., Cheng, A., Bradlow, H., Bradlow, S., Carragher, B., and Potter, C.S. (2008a). A test-bed for optimizing high-resolution single particle reconstructions. J Struct Biol 163, 29-39.
Stagg, S.M., LaPointe, P., and Balch, W.E. (2007). Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol 17, 221-228.
Stagg, S.M., LaPointe, P., Razvi, A., Gurkan, C., Potter, C.S., Carragher, B., and Balch, W.E. (2008b). Structural basis for cargo regulation of COPII coat assembly. Cell 134, 474-484.
Strable, E., Prasuhn, D.E., Jr., Udit, A.K., Brown, S., Link, A.J., Ngo, J.T., Lander, G., Quispe, J., Potter, C.S., Carragher, B., et al. (2008). Unnatural amino acid incorporation into virus-like particles. Bioconjug Chem 19, 866-875.
Voss, N.R., Lyumkis, D., Cheng, A., Lau, P.W., Mulder, A., Lander, G.C., Brignole, E.J., Fellmann, D., Irving, C., Jacovetty, E.L., et al. (2010). A toolbox for ab initio 3-D reconstructions in single-particle electron microscopy. J Struct Biol 169, 389-398.
Xu, Y., Song, S., Moeller, A., Kim, N., Piao, S., Sim, S.H., Kang, M., Yu, W., Cho, H.S., Chang, I., et al. (2011). Functional implications of an intermeshing cogwheel-like interaction between TolC and MacA in the action of macrolide-specific efflux pump MacAB-TolC. J Biol Chem.